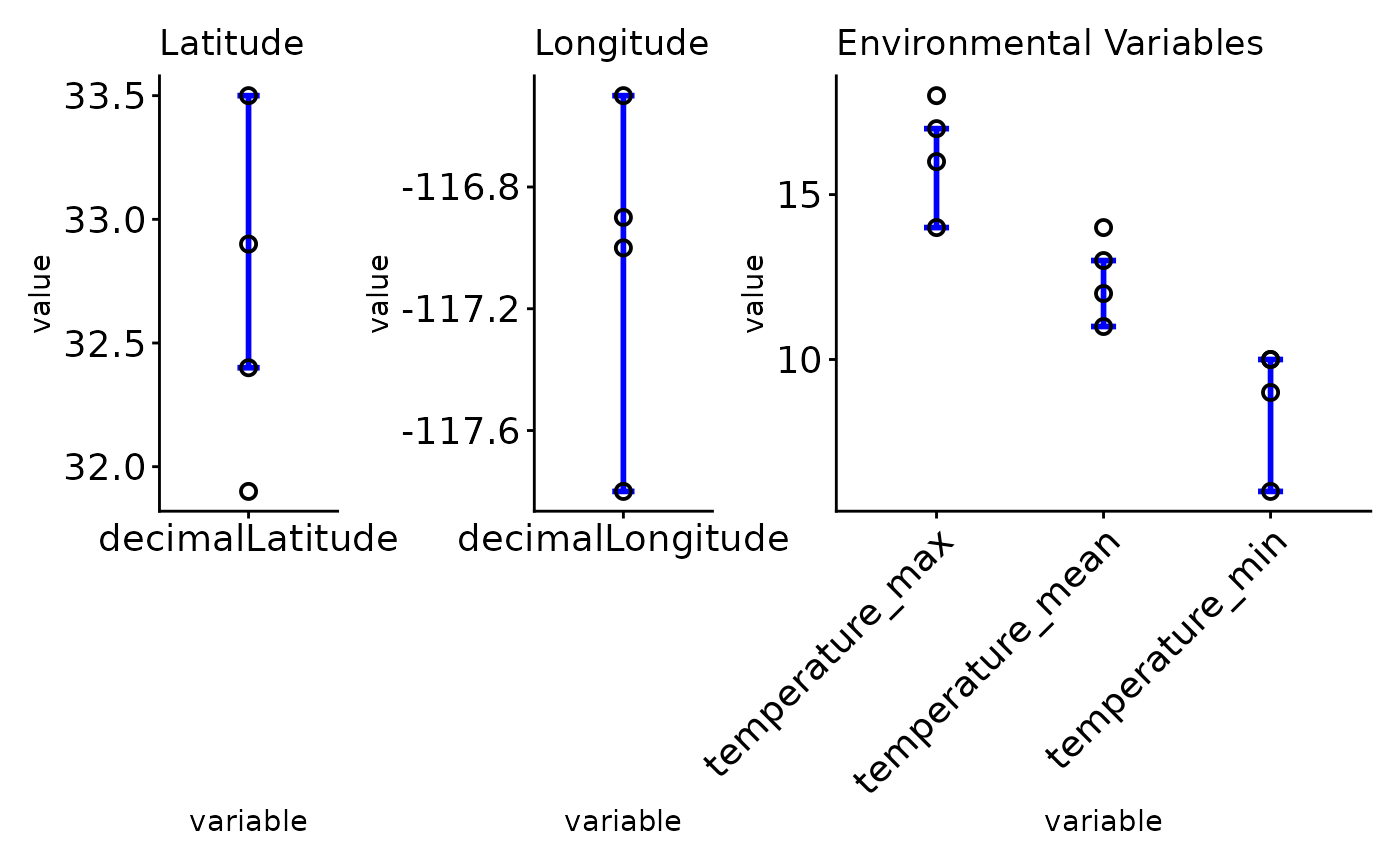
Plot cleaned data overlay overall occurrence data to demonstrate accepted ranges of spatial and non-spatial attributes
ec_plot_var_range.RdPlot cleaned data overlay overall occurrence data to demonstrate accepted ranges of spatial and non-spatial attributes
Usage
ec_plot_var_range(
data,
summary_df,
latitude = "decimalLatitude",
longitude = "decimalLongitude",
env_layers
)Value
A plot which shows spatial and environmental variables with the acceptable range for species habitability
Examples
data <- data.frame(
scientificName = "Mexacanthina lugubris",
decimalLongitude = c(-117, -117.8, -116.9, -116.5),
decimalLatitude = c(32.9, 33.5, 31.9, 32.4),
temperature_mean = c(12, 13, 14, 11),
temperature_min = c(9, 6, 10, 10),
temperature_max = c(14, 16, 18, 17),
flag_outlier = c(0, 0.5, 1, 0.7)
) # this data table has data points which was considered as outliers
data_x <- data.frame(
scientificName = "Mexacanthina lugubris",
decimalLongitude = c(-117, -117.8, -116.5),
decimalLatitude = c(32.9, 33.5, 32.4),
temperature_mean = c(12, 13, 11),
temperature_min = c(9, 6, 10),
temperature_max = c(14, 16, 17),
flag_outlier = c(0, 0.5, 0.7)
)
# cleaned data base after removing outliers >x probability.
# in this example, removed data points >0.7 probability to be
# considering outliers
env_layers <- c("temperature_mean", "temperature_min", "temperature_max")
summary_df <- ec_var_summary(data_x,
latitude = "decimalLatitude",
longitude = "decimalLongitude",
env_layers
)
# this is the final cleaned data table which
# will be used to derive summary of acceptable niche
ec_plot_var_range(data,
summary_df,
latitude = "decimalLatitude",
longitude = "decimalLongitude",
env_layers
)